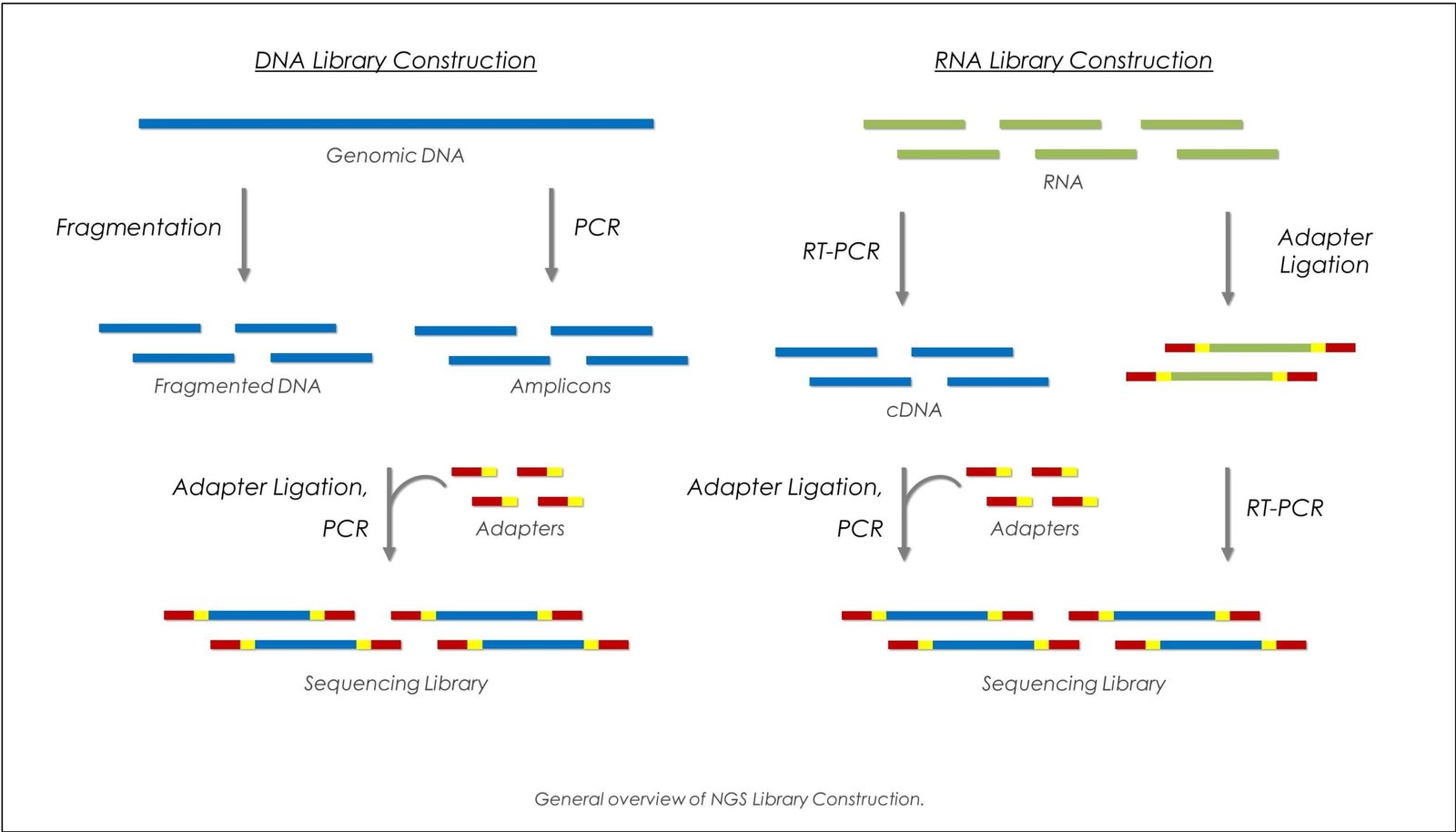
The next-generation sequencing (NGS) has revolutionized the approach to genomes, transcriptomes, and epigenetic alterations. Quality library preparation to the conversion of nucleic acids into forms that can be sequenced is the key to successful NGS project. There are multiple approaches to it, and every one of them has definite merits and drawbacks that you are to take into consideration prior to choosing one of them to implement in your study, and here we may examine five key points to keep in consideration when choosing your approach about the research objectives.
Sample Input Requirement and Quality
The quantity and quality of starting material is a basic consideration in selecting a library preparation method. Other procedures are meant to work with high-input samples, in which case enough, high-quality DNA or RNA exists. Conversely, other protocols like segmentation based protocols are able to process low-input samples with less degradations. As an example, to perform in traditional ligation-based approaches, the amount of input DNA can be microgram scale, but approaches such as Nextera involve using other enzymes, such as transposase that results in DNA fragmentation and addition of unique barcodes, allowing library preparation to nanogram quantities of DNA.
In case your work implies working with clinical samples, ancient DNA, and single-cell sequencing, remember to use low input/degraded sample protocols. It is important to balance on the sensitivity and protocol amplification bias, the low-input protocols usually involve more cycles of PCR which is available to generate errors or bias the representation version of these regions. Knowing the quality and quantity of your samples are the initial phases towards selecting a protocol that could optimize the effectiveness level of your sequencing run.
Turnaround time and Workflow complexity
The protocols to prepare NGS libraries differ radically by factors of complexity of the workflow, wet time, and turnaround. The automated and streamlined workflows are the requirement of high-throughput labs, and sometimes more time- and effort-consuming multi-enzymatic reactions or other purification steps used in the case of the conventional methods can be replaced with a single reaction of tagmentation in the case of tagmentation-based approaches, which reduces preparation time and even the risk of sample loss.
The ideal NGS library preparation services should assist you to streamline your routine by inculcating quality control and automation, so that even a complicated protocol is taken through, and delivers high quality data efficiently.
When choosing a protocol, consider the capacity of your laboratory and schedule. The simpler project workflow may be suitable to the project with a strict deadline or high sample volume and the researchers desire more consistent library quality whereas the more hands-on approach can be more suitable to the deeper customization needs or researchers working with difficult samples. However, the approach that you choose to adopt at the end of the day must accommodate throughput needs on top of your technical involvement requirements.
Library Complexity, Coverage and Bias
Every NGS experiment is aimed at producing the quality data that is representative of the genomic content. Coverage bias may be evident because of library preparation through which complexity can be skewed. As an example, PCR amplification may be required when beginning with low amounts of input, but itself can bias the representation of particular sequences, thus over- or under-representing regions with high or low GC content. This bias is minimized by PCR-free methods, wherever practicable, and frequently leads to more uniform genome coverage.
The trade-off possibilities between yield and bias need to be evaluated. Although PCR-based methods may be advantageous in amplifying low-input samples, there is a chance they may decrease library complexity resulting in a possible difficulty in the visualization of some rare variants. Moreover, some of the fragmentation methods, e.g. enzymatic fragmentation or sonication can result in the fragments of varying end sequences which have an impact on the sequencing downstream. Use the biases present in the protocol as a guide to determining the protocol to adapt to your experimental goals the putative comprehensiveness of genome coverage, sensitivity to low-frequency mutations, or emphasis on selected regions of the genome.
Scalabilty and Price Factor
The budget limitations will always be a part of research and the costs of NGS library preparation methods differ significantly. The choice of which method to use could frequently be based not merely on initial costs of reagents and consumables but scalability into higher throughput protocols; segmentation based methods are relatively low-cost in high-throughput applications owing to the simplified workflow and smaller reagent volumes; in contrast custom protocols optimized to have high fidelity and lower bias may be more expensive due to the increased clean-up or quality controls steps performed in custom protocols designed to have higher fidelity/lesser bias.
You should also think on the scalability of your approach in general. Compatibility with automation can dramatically reduce labor costs in large scale studies or core facilities and increase reproducibility. The costs of the samples can be further minimized by investing in methods that allow parallel processing without sacrificing quality, and this too it is important that the tradeoffs between cost and performance are viewed through the lens of your research budget and planned scope of operation.
Application-Specific Considerations
Last but not least is the choice of NGS library preparation that is based on research questions you have. Various applications – such as whole genome sequencing, targeted sequencing, RNA-Seq library preparation or epigenetic studies – have different needs; e.g. RNA-Seq library preparation needs to preserve strand identity whilst considering the relative abundance of different transcripts whereas methods designed to study methylation may require additional steps such as bisulfite conversion.
There are also those that have been designed to be suitable in applications. An example would be that the single cell RNA-seq platforms make use of advanced library prep kits which in turn allow thousands of individual cells at a single time to be captured and barcoded. The parameters of the experiment provided by researchers including sample type, desired resolution level and downstream requirements of analysis should be carefully taken into consideration so that an RNA-Seq protocol with the best sensitivity-specificity-data trade off could be chosen.
Conclusion
When you choose a method of NGS library preparation, you should consider many factors, such as input sample, efficiency of the workflow, quality of data, cost, and the objectives of your experiment. By giving special attention to these five most important details including sample requirements, workflow complexity, library bias/coverage requirements/cost concerns besides certain application-specific requirements; you can choose the best option that would allow maximizing the sequencing outcome. An appropriate approach does not only fall within technical and budgetary considerations but also accelerates your research ahead by giving you strong and reproducible results.
On the one hand, it is important to be aware of the changes that are sweeping NGS technologies because that keeps you current on library preparation, and on the other hand, it is important to be up to date via literature, and workshops, and liaising with specialists in the field to perfect your methods and to get the maximum out of what NGS can offer. Protected planning and the appropriate choice of method will allow it to see the light and make its contribution towards innovation and discovery.